Operations: Difference between revisions
Tlauritsen (talk | contribs) No edit summary |
Tlauritsen (talk | contribs) |
||
| (118 intermediate revisions by 6 users not shown) | |||
| Line 1: | Line 1: | ||
==To Start the DAQ under VNC== | |||
instructions 3/6/2021 (from Claus) | |||
on WS3, from gretina account, start the VNC | |||
server and make a local VNC viewer | |||
vncserver -list should give you a list of all running sessions | |||
vncserver -kill :3 will kill the running session as it may be broken | |||
rm /tmp/.X11-unix/X3 delete a hidden file, if the kill command doesn't work | |||
vncserver :3 start a new session | |||
vncviewer :3 opens the viewer locally, you can also follow instructions to connect remotely | |||
now you should do these things in the VNC | |||
(which is local on WS3): | |||
ssh -X a2 | |||
xset fp+ /usr/share/X11/fonts/75dpi fixes a font problem, otherwise EPICS control will not start | |||
./startGRETINA | |||
that should bring some of the GT DAQ up. | |||
Now setup the DAQ | |||
kill DAQ | |||
initialize DAQ | |||
GRETINA+FMA | |||
now the runcontrol should come up and you | |||
can start a run. Note: now you have to start and | |||
stop runs from a terminal as: | |||
gDAQon | |||
gDAQoff | |||
It increments the run number itself. | |||
next | |||
firefox | |||
connect to the data room blue jeans | |||
share your screen | |||
Now people on the dataroom BJ can see what you | |||
see without having to make their own VNC connection | |||
If you are also running the DFMA DAQ, you should follow the | |||
instructions below to get that DAQ going as well. | |||
In a terminal window (5) | |||
ssh -X gretina@dmfa_darek | |||
./dfmaCSS | |||
check all IOCs are up or cycle power | |||
get to trigger IOC and reboot | |||
on DFMACommander.opi | |||
scripts | |||
setup_system_sync | |||
now DFMA should be up; but not synced to GT yet. To do that | |||
got to GT DAQ | |||
stop run | |||
Syncro FIX | |||
start run | |||
Now, in a terminalwindow (4): | |||
ssh -X gretina@dmfa_darek | |||
cd /global/data1a/gretina/exp1861_Lotay_data1a/dfmadata | |||
here you can | |||
start_run.sh xxx | |||
stop_run.sh | |||
being in sync with the GT DAQ regarding the run numbers | |||
==To Start the DAQ == | |||
[[File:grt1.jpg|right]] | |||
[historical entry] | |||
issue this command to get the fonts necessary for EDM | |||
* xset fp+ /usr/share/X11/fonts/75dpi | |||
* There are two ways to start the Gretina data acquisition system, depending on the computer in use; | |||
# On the iMac: Double click on the Gretina icon (start_DAQ) located on the top left corner of the iMac's desktop and the window (shown to the right --->) will pop up. | |||
# On the WS2 and WS3 Linux machines: First logon to a1.gam or a2.gam using the commands below | |||
:: ssh -X [email protected] | |||
:: or | |||
:: ssh -X [email protected] | |||
Once in the home directory ~/, type the command below to bring up the Gretina DAQ GUI (shown to the right --->). | |||
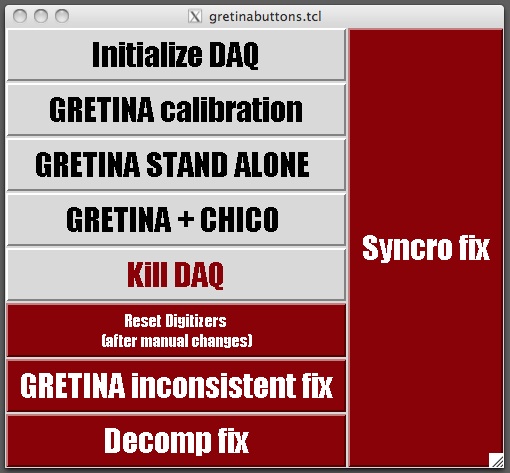
./gretinabuttons.tcl | |||
{| cellpadding=5 style="border:1px solid #AA6633" | |||
|- bgcolor="#fafeff" | |||
| To make sure the DAQ is not running anywhere else, first PRESS | |||
|} | |||
KILL DAQ | |||
Wait until you see in the lower shell: stop. Everything should close now. Then PRESS | |||
INITIALISE DAQ | |||
{| cellpadding=5 style="border:1px solid #AA6633" | |||
|- bgcolor="#fafeff" | |||
| This will take a few moments, up to 1-2 minutes. Observe the lower shell and check that it is finally stating: READY FOR CALIBRATION | |||
|} | |||
In this mode, GRETINA is ready for data taking in calibration mode, meaning: validation mode, 0.2us traces. Please start a run, if you just want to make a calibration. | |||
If you want to couple GRETINA to CHICO [or run it standalone] in DECOMP mode, press either | |||
GRETINA STAND ALONE | |||
or | |||
GRETINA+CHICO | |||
Again, things will happen in the shell and it is loading bases and setting up the triggers. | |||
NOTE: the only difference between GRETINA STAND ALONE and GRETINA+CHICO is the trigger mask. In order to enable an external trigger and to disable the internal triggers, the ROUTERS will be changed correspondingly. | |||
{| cellpadding=5 style="border:1px solid #AA6633" | |||
|- bgcolor="#fafeff" | |||
| Now: GRETINA is set up to the correct mode and the CLUSTER STATE [in CLUSTER → NODES] should be ok. If not, please wait for a certain amount of time [could be more than 5 minutes]. If this does not help: try the full procedure again. | |||
|} | |||
You may want to go to the Cluster page and select the proper data | |||
format, either 'mode2/mode3' or just 'mode2'. The default seems | |||
to be 'mode2/mode3'. Note: you should do that before you take any data. | |||
==To Start the DAQ (old way)== | |||
[historical entry] | |||
* We run the DAQ from "a1.gam" (for now) | |||
ssh -X [email protected] | |||
or | |||
ssh -X [email protected] | |||
* you may have to issue this command to get access to the proper fonts for the EPICS displays | |||
xset fp+ /usr/share/X11/fonts/75dpi | |||
* Go to the gretina home directory | |||
cd | |||
source goatlas | |||
* The MENU below should show up: | |||
---------- MENU ---------- | |||
VME 1 | |||
Display 2 | |||
Run Control 3 | |||
Run ControlBGS 3b | |||
Soft IOC 4 | |||
Soft IOC2 4b | |||
CAEN PS 5 | |||
Cluster 6 | |||
Cluster Kill 8 | |||
Mode 2 9 | |||
Mode 3 10 | |||
Mode 4 11 | |||
RESET daq InBeam 12 | |||
RESET daq Gamma 12b | |||
StripTool 13 | |||
Calibrate 14 | |||
EXIT E | |||
KILL ALL K | |||
-------------------------- | |||
* Info: Exiting the MENU does not exit the DAQ. One can return to the MENU anytime by typing $PWD/godaq | |||
* Start things in the following order: | |||
K, 4, 4b, 2, 3, 6 | |||
K to make sure something is not running somewhere. After this sequence, verify you see "CA Connected" (CA: Channel Access) in the RunControl panel | |||
* Press Button "cluster" in the GlobalControl panel to chose the correct data mode (mode 2: decomposed data, mode 3: raw data (traces)). If GT is not set up already for mode2 data taking (producing decomposed data) you should invoke option 9. | |||
Note: In the Gretina RunControl panel, 'new dir' makes the run starts at 0002, switching back to an existing directory makes the run start at the previous Run number + 2 | |||
* Press button "global control" | |||
The upcoming panel is being used for setting global | |||
parameters for ALL chrystals (baseline, polarity, etc.) | |||
Raw delay: For internal mode data: 3.7 usec (for analyzing the traces) | |||
typically you will see 2.7 usec here | |||
Raw len: length of the trace in usec | |||
don't go below 0.2 usec (corresponding to 20 | |||
short words, while the header is already 16 words long, | |||
so there are 4 words left for the trace itself) | |||
in decomposition mode: Use 2.0 usec | |||
(corresponding to 16 words for the header and 184 | |||
words for the trace) | |||
Threshold setting: typically cc: 1000, segment 300 (do not go below 300 | |||
in the segments and 150 in CC) | |||
in "user trigger" you can set the crystal readout rate up to 20,000. | |||
Lower this number if the decomposition cluster cannot keep up | |||
* Press button "TimeStamps" in the Gretina DAQ top level to check if the timestamps are still synchronized imp sync: starts a train of imperative sync signals, by pressing the button again, the train stops single pulse imp sync: One single imperative sync pulse | |||
* press button "pseudoScalers" in the Gretina DAQ top level to verify that data is coming in and being processed | |||
* rebooting digitizer IOCs | |||
ssh gsoper:[email protected] | |||
find the number in the "timestamp window". simply count to | |||
get the xx value to use. For crate 3, use 03. Do not ssh from | |||
the terminal you set up GT from as you will need to type "~." | |||
to get out and that will kill your window as well. Always ssh | |||
from a seperate terminal window | |||
if you telnet into the IOC and you see tasks suspended (type i | |||
to see the processes and thir status) you need to reboot. Simply | |||
type "reboot". Using "tr" to start them usually does not help | |||
after a reboot: push the "reset after an IOC reboot" butten | |||
and the "TTCS settings button" and the other buttons there. | |||
for access to the trigger IOC, | |||
ssh gsoper:[email protected] | |||
Once all is set up, you can also start and stop runs on the command line with these commands (scripts in ~/scripts) | |||
gDAQon (does not work?) | |||
gDAQoff (works) | |||
The alarm page is accessed through the GT main DAQ screen upper right corner. | |||
This is where you can mute the alarm | |||
==to stop the DAQ== | ==to stop the DAQ== | ||
== | push the stop button in run control window | ||
If you want to stop everything, select "K" in | |||
the $PWD/godaq script. | |||
==to shut down the DAQ== | |||
* Type 8 in the MENU to kill the Cluster | |||
* Type K (Kill All) in the MENU to kill the rest | |||
==online sorting with GEBSort, to a map file== | |||
NOTE: you can only receive on-line data if the decomposition cluster is running. The Global Event Builder, where GEBSort gets it data from, is only active if the decomposition cluster is up and running | |||
cd workdir (e.g., | Here is an example of running GEBSort on ws3.gam taking data from the GEB | ||
to a map file we can look at while we take data | |||
o ssh -X [email protected] | |||
o Download [THIS IS OUT OF DATE] | |||
cd workdir (e.g., /home/gtuser/gebsort) | |||
https://svn.anl.gov/repos/gs_analysis/GEBSort . | |||
or | or | ||
wget http://www.phy.anl.gov/gretina/GEBSort/AAAtar.tgz | wget http://www.phy.anl.gov/gretina/GEBSort/AAAtar.tgz | ||
tar -zxvf AAAtar.tgz | tar -zxvf AAAtar.tgz | ||
on a1.gam you would need to do this as well | |||
export ROOTSYS=/home/users/tl/root/root-v5-34-00 | export ROOTSYS=/home/users/tl/root/root-v5-34-00 | ||
export PATH=$ROOTSYS/bin:$PATH | export PATH=$ROOTSYS/bin:$PATH | ||
export LD_LIBRARY_PATH=/home/users/tl/root/root-v5-34-00/lib/root | export LD_LIBRARY_PATH=/home/users/tl/root/root-v5-34-00/lib/root | ||
on ws3 or ws2 you should be fine | |||
o now compile everything | |||
rm curEPICS; ln -s /global/devel/base/R3.14.11 curEPICS | |||
make clean | |||
make | |||
make GEBSort | make GEBSort | ||
./mkMap > map.dat | |||
rm GTDATA; ln -s ./ GTDATA | |||
o to run and take data, type | |||
# +-- Global Event Builder (GEB) host IP (or simulator) | |||
# | + -- Number of events asked for on each read | |||
# | | +-- desired data type (0 is all) | |||
# | | | +-- timeout (sec) | |||
# | | | | | |||
./GEBSort \ | |||
-input geb 10.0.1.100 100 0 100.0 \ | |||
-mapfile c1.map 200000000 0x9ef6e000 \ | |||
-chat GEBSort.chat | |||
you may also just say | |||
go geb_map | |||
o to display the data in the map file | |||
rootn.exe -l | |||
compile GSUtils (first line) | |||
sload c1.map file (third line) | |||
update (fourth line and other places) | |||
display | |||
o note: It can take a while, up to a minute or so, before the GEB has data to offer to ./GEBSort so if ./GEBSort doesn't get data, just try again after you have started a run. | |||
==off-line sorting with GEBSort, to a root file== | |||
Here is an example of running GEBSort with output to a root file in an off-line situation | |||
o Download the software | |||
cd workdir (e.g., /home/gtuser/gebsort) | |||
git clone https://gitlab.phy.anl.gov/tlauritsen/GEBSort.git . | |||
o now compile everything | |||
make clean | |||
make | |||
./mkMap > map.dat | |||
rm GTDATA; ln -s ./ GTDATA | |||
Ignore errors about not being able to make 'GEBSort' if you don't have VxWorks and EPICS installed! It will make 'GEBSort_nogeb', which is the version of GEBSort we will use for off-line data sorting where VxWorks and EPICS are not needed (the on-line situation, where you DO need GEBSort, is described above). | |||
o sort your (merged) file as: | |||
./GEBSort_nogeb \ | |||
-input disk merged_data.gtd \ | |||
-rootfile test.root RECREATE \ | |||
-chat GEBSort.chat | |||
note1: for DGS data, GEBSort_nogeb works equally well on the idividual files from gtReceiver4 as the merge of those files created with GEBMerge | |||
o to display the data in the root file | |||
root-l | rootn.exe -l (or 'root -l' since it is just a root file) | ||
compile GSUtils | compile GSUtils (first line) | ||
dload test.root file (third line) | |||
display | display | ||
In general, the 'go' file of the GEBSort package contains a lot of examples. You can make your own script file to process your data using these examples. | |||
==FAQ== | ==FAQ== | ||
* | ==misc problem fixes and procedures== | ||
* To fix a non-responsive timestamp, i.e. just counts and never resets, use command "caput Cry[#]_CS_Fix (e.g., caput Cry4_CS_Fix 1) 1" where [#] is the bank # of the offending timestamp(s). This will reload the FPGA code and reboot the IOC. Any time an IOC is rebooted, you should press the "RESET after an IOC Reboot" button on Global Controls edm screen. Then choose the settings type you want, usually TTCS or Validate settings, on the same screen. Note: this only works if the IOC is still respnding to CA. If it doesn't, try access through the terminal server and reboot, or cycle the power on the crate. Typically you have to manually reboot when tExcTask and tNetTask are suspended. | |||
* to select the decomposition nodes that should be runnin (and, thus, avoid any nodes that are broken), edit the files: "setup7modulesRaw.sh", "setupEndToEnd28.sh" in the directory: "/global/devel/gretTop/11-1/gretClust/bin/linux-x86". Then do 8 to kill , then run the scripts you just changed, and 6 to bring up decomposition cluster up again. On dogs, type 'wwtop', to see details of what nodes are running. | |||
* to bring up the GT repair buttons: "~/gretinabuttons.tcl". you may have to do that from a1.gam for now as we have a problem doing some things from the linux WS machine; | |||
* If some crystals are not getteing decomposed or running properly: stop, do imp sync, and start again. If that is not enough, also click on the 4 buttons in the south east corner of the "global Control" page. If it still does not decompose, stop the decomposition cluster and start it again in the main DAQ script | |||
* set the buffer size in the GEB, use | |||
caput GEB_CS_Delay 5 | |||
in this case, we set it to 5 seconds. 5 to 10 are good values. | |||
* another mysterious thing to try | |||
LEDAT 19 700;LEDAT 23 800;SetPZMultCC.sh;SETLEDWIN 30 | |||
== Calibration Procedure ANL -- version == | |||
===get the data=== | |||
To obtain the data needed to do the segment and CC calibration, we change the GT configuration to | |||
Raw Len = 0.20 us | |||
Trig Mode = Validate | |||
mode 3 data (in godaq script) | |||
the cluster nodes should be running 1 sendRaw process per crystal | |||
in cluster, select 'mode 2 and 3' | |||
you may have to adjust 'Crystal readout Rate Limit' to adjust the rate. In this mode you can take data at a rate of 60 times faster than when you run your data through the decomposition cluster to obtain mode2 data. You can split the files using this handy utility | |||
gMultiRun <number of runs> <minutes per run> <minutes between runs> | |||
check that the | |||
GlobalRaw.dat | |||
file increments. There is also a Global.dat, but it should be empty. | |||
===run the calibration software=== | |||
Shaofei is working on this software. Eventually we will add it to GEBSort, where the date will be processed in the 'bin_gtcal' module. | |||
== Calibration Procedure -- NSCL version == | |||
NOTE: these instructions were lifted from NSCL, we will have to modify them to the needs at ANL | |||
In broad strokes, the procedure has been to fit source spectra for the central contacts (CC), and then calibrate the segments based on the observed energy in a segment vs. the calibrated CC energy for multiplicity 1 events. There are some simple, and fairly quick unpacking codes which make the necessary ROOT spectra, and then some ROOT macros that actually perform the fitting etc. The procedure is outlined below, and the codes are all located in /home/users/gretina/Calibration/. | |||
=== CC Calibration === | |||
1. Unpack the calibration data (this all assumes it's mode3 data) using the code ./UnpackRaw, where the commands are | |||
./UnpackRaw <Data Directory, i.e. MSUJune2012> <Run#, i.e. 0061> | |||
2. Run the macro to fit the peaks: | |||
root -l | |||
root [0] .L FitSpectra.C | |||
root [1] FitCCSpectra(.root Filename, i.e. "MSUMay2012Run0060.root", | |||
value for xmin, typically choose 100, | |||
Boolean for manual checking, to check, set to 1, otherwise 0, | |||
source type, i.e. "Eu152"); | |||
-- Here the inputs are the .root filename (i.e. "MSUMay2012Run0061.root") which was generated in the unpacking step, a minimum spectrum value to look at (not so important, just pick something low, like 100 or something), a boolean to say if you want to personally approve the fits (CHECK = 1), or just let it go (CHECK = 0), and the source type (options are "Co60", "Bi207", "Eu152", "Co56" and "Ra226" right now). | |||
-- If you run this script with CHECK == 1, you run in a quasi-interactive mode. That is, for each CC, you'll get a chance to look at all of the fits, and to move to the next CC, you must accept the fits by hitting 'y', and then enter. If you see one you don't like, you can enter the peak number, and the fit will be redone, with tweaked parameters. Then you can either accept the peak, or reject it, in which case it's ignored in fitting the calibration. Peaks are numbered in order of increasing energy, and labeled in the ROOT canvas, so you know which number to type for a given fit. The only thing to note in this is that if peaks are < 20 keV apart, they are fit together. In this case, if peaks 7 + 8 are fit together, there will only be a spectrum shown in the position corresponding to peak 7. If you're unhappy with the fit though, and refit 7, it will only fit peak 7. If you want peak 8 retried as well, you need to specify this additionally. | |||
-- Regardless of the value of CHECK, if the code can't find a peak, it'll ask you if you see one. If you do, you can enter the approximate channel number, and a fit will be attempted. If you don't, entering -1 will just leave out the calibration point. | |||
-- Output from the macro: | |||
paramCC<ROOTFilename>.dat (i.e. paramCCMSUMay2012Run0061.dat) -- contains all fit parameters | |||
meanCC<ROOTFilename>.dat (i.e. meanCCMSUMay2012Run0061.dat) -- contains just the centroid of the fits | |||
3. After you have fit all the source spectra you're interested in, you can fit the calibration with the ROOT macro: | |||
root -l | |||
root [0] .L FitCalibration.C | |||
root [1] FitCalibration(number of calibration files, i.e. 3, | |||
First .root file, i.e. "MSUMay2012Run0060.root", | |||
Second .root file, i.e. "MSUMay2012Run0071.root", | |||
Third .root file, i.e. "MSUMay2012Run0070.root", | |||
Fourth .root file, i.e. "" if only using three files, | |||
Flag for CCONLY = 1 if only fitting CC, | |||
Flag for making detector maps, DETMAPS = 0 for NO); | |||
-- This literally fits the calibration points with a straight line, and calculates the residuals, etc. of the fit. Input to the macro is the number of files you want to use in the calibration (number), the .root filenames (i.e. "MSUMay2012Run0061.root"), a boolean to say if you only want to fit the CC (answer should be yes, so CCONLY = 1), and a boolean to see if you want to make the detector map files for decomposition (at this stage with only CC calibrations, no). Note, if you use less than 4 calibration files, for the unused filenames, just put "". | |||
-- Output from the macro: | |||
fitFile<ROOTFilename#1>.root | |||
(i.e. fitFileMSUMay2012Run0061.root) | |||
-- contains the calibration fit, and residual plots | |||
finalCalOutput<ROOTFilename#1>.dat | |||
(i.e. finalCalOutputMSUMay2012Run0061.dat) | |||
-- contains three columns - ID, offset and slope | |||
This is the file I rename (EhiGainCor-CC-MMDDYYYY.dat) and use for CC calibration in unpacking codes. | |||
finalDiffOutput<ROOTFilename#1>.dat (i.e. finalDiffOutputMSUMay2012Run0061.dat) | |||
-- contains residual information in text format | |||
=== Segment Calibration === | |||
1. Edit the unpack-m1.C source code (in the subdirectory src/), and change the line that reads in the calibration to read the new CC calibration that you've created. | |||
RdGeCalFile("<Name of CC calibration - i.e. finalFitFile<>.dat>", ehiGeOffset, ehiGeGain); | |||
2. Remake the code, just type make. This replaces the executable in the directory above. | |||
3. Run the code UnpackM1 on the data you're using for the segment calibration. Typically, pick something with statistics enough to see something in the back segments. | |||
./UnpackMult1 <Directory, i.e. MSUMay2012> <Run Number, i.e. 0061> | |||
-- This code takes a while, because it scans the file 28 times, to calibrate each crystal one at a time. This is because it's using 2D ROOT matrices, and I can't make the memory usage work properly to do multiple crystals at the same time. It always has a memory allocation problem. If someone knows how to fix this, by all means, please do. Let Heather know how you fix it too =) | |||
-- The code is essentially sorting segment multiplicity 1 events, and using the calibrated CC to calibrate the segment energies. Right now, it's looking at the 5MeV CC to do this. You can change this in the code, if desired. At the end of the code, it then fits the plot of CC vs. segE, and extracts a slope and offset. This is actually then written out as a completed calibration file (see output information). | |||
-- Output files from code: | |||
<ROOTFilename>Crystal##.root | |||
where ## goes from 1 to 28 -- these are .root files | |||
containing the 2D plots for each crystal, as well as projections | |||
<ROOTFilename>.slope | |||
-- contains channel ID (electronics ID), offset and slopes | |||
-- the finished calibration | |||
This is the file that is usually renamed to | |||
EhiGainCor-MMDDYYY.dat and used in codes. | |||
=== Detector Map and Trace Gain Generation for Decomp === | |||
1. Edit the ROOT macro file FitCalibration.C, specifically the function MakeDetectorMaps(), to read in the appropriate calibration file. Also edit the destination directory for the generated detector map files, which appears in a few places... | |||
calibrationFile = fopen("<CalibrationFilename.slope>", "r"); | |||
... | |||
system("mkdir <DestinationDirectoryName>"); | |||
... | |||
detmap_file = fopen(Form(<DestinationDirectoryName>/detmap_Q%dpos%d_CC%d.txt", quad, position, ccnum), "w"); | |||
... | |||
tracegainfile = fopen(Form(<DestinationDirectoryName>/tr_gain_Q%dpos%d_CC%d.txt", quad, position, ccnum), "w"); | |||
2. Run the MakeDetectorMaps() macro. | |||
root -l | |||
root[0] .L FitCalibration.C | |||
root[1] MakeDetectorMaps() | |||
3. Detector map and trace gain files should be automatically generated in the directory specified within the macro. | |||
* | === Statistics required for calibration === | ||
* 56Co, mode 3, validate, trace 0.2us --- 8 GB = ~15 minutes | |||
* 152Eu, mode 3, validate, trace 0.2us --- 6 GB = ~15 minutes | |||
* 226Ra, mode 3, validate, trace 0.2us --- 8 GB = ~15 minutes | |||
=== Statistics required for checking segments resolution === | |||
* 60Co (10 uC), mode 3, validate, trace 0.2us --- 30 GB = ~60 minutes --- 500 cts per segments of the rear layer | |||
Latest revision as of 21:07, November 18, 2022
To Start the DAQ under VNC
instructions 3/6/2021 (from Claus)
on WS3, from gretina account, start the VNC server and make a local VNC viewer
vncserver -list should give you a list of all running sessions vncserver -kill :3 will kill the running session as it may be broken rm /tmp/.X11-unix/X3 delete a hidden file, if the kill command doesn't work vncserver :3 start a new session vncviewer :3 opens the viewer locally, you can also follow instructions to connect remotely
now you should do these things in the VNC (which is local on WS3):
ssh -X a2 xset fp+ /usr/share/X11/fonts/75dpi fixes a font problem, otherwise EPICS control will not start ./startGRETINA
that should bring some of the GT DAQ up. Now setup the DAQ
kill DAQ initialize DAQ GRETINA+FMA
now the runcontrol should come up and you can start a run. Note: now you have to start and stop runs from a terminal as:
gDAQon gDAQoff
It increments the run number itself.
next
firefox connect to the data room blue jeans share your screen
Now people on the dataroom BJ can see what you see without having to make their own VNC connection
If you are also running the DFMA DAQ, you should follow the instructions below to get that DAQ going as well.
In a terminal window (5)
ssh -X gretina@dmfa_darek ./dfmaCSS check all IOCs are up or cycle power get to trigger IOC and reboot on DFMACommander.opi scripts setup_system_sync
now DFMA should be up; but not synced to GT yet. To do that
got to GT DAQ stop run Syncro FIX start run
Now, in a terminalwindow (4):
ssh -X gretina@dmfa_darek cd /global/data1a/gretina/exp1861_Lotay_data1a/dfmadata
here you can
start_run.sh xxx stop_run.sh
being in sync with the GT DAQ regarding the run numbers
To Start the DAQ
[historical entry]
issue this command to get the fonts necessary for EDM
- xset fp+ /usr/share/X11/fonts/75dpi
- There are two ways to start the Gretina data acquisition system, depending on the computer in use;
- On the iMac: Double click on the Gretina icon (start_DAQ) located on the top left corner of the iMac's desktop and the window (shown to the right --->) will pop up.
- On the WS2 and WS3 Linux machines: First logon to a1.gam or a2.gam using the commands below
- ssh -X [email protected]
- or
- ssh -X [email protected]
Once in the home directory ~/, type the command below to bring up the Gretina DAQ GUI (shown to the right --->).
./gretinabuttons.tcl
| To make sure the DAQ is not running anywhere else, first PRESS |
KILL DAQ
Wait until you see in the lower shell: stop. Everything should close now. Then PRESS
INITIALISE DAQ
| This will take a few moments, up to 1-2 minutes. Observe the lower shell and check that it is finally stating: READY FOR CALIBRATION |
In this mode, GRETINA is ready for data taking in calibration mode, meaning: validation mode, 0.2us traces. Please start a run, if you just want to make a calibration.
If you want to couple GRETINA to CHICO [or run it standalone] in DECOMP mode, press either
GRETINA STAND ALONE
or
GRETINA+CHICO
Again, things will happen in the shell and it is loading bases and setting up the triggers.
NOTE: the only difference between GRETINA STAND ALONE and GRETINA+CHICO is the trigger mask. In order to enable an external trigger and to disable the internal triggers, the ROUTERS will be changed correspondingly.
| Now: GRETINA is set up to the correct mode and the CLUSTER STATE [in CLUSTER → NODES] should be ok. If not, please wait for a certain amount of time [could be more than 5 minutes]. If this does not help: try the full procedure again. |
You may want to go to the Cluster page and select the proper data format, either 'mode2/mode3' or just 'mode2'. The default seems to be 'mode2/mode3'. Note: you should do that before you take any data.
To Start the DAQ (old way)
[historical entry]
- We run the DAQ from "a1.gam" (for now)
ssh -X [email protected] or ssh -X [email protected]
- you may have to issue this command to get access to the proper fonts for the EPICS displays
xset fp+ /usr/share/X11/fonts/75dpi
- Go to the gretina home directory
cd source goatlas
- The MENU below should show up:
---------- MENU ---------- VME 1 Display 2 Run Control 3 Run ControlBGS 3b Soft IOC 4 Soft IOC2 4b CAEN PS 5 Cluster 6 Cluster Kill 8 Mode 2 9 Mode 3 10 Mode 4 11 RESET daq InBeam 12 RESET daq Gamma 12b StripTool 13 Calibrate 14 EXIT E KILL ALL K
- Info: Exiting the MENU does not exit the DAQ. One can return to the MENU anytime by typing $PWD/godaq
- Start things in the following order:
K, 4, 4b, 2, 3, 6
K to make sure something is not running somewhere. After this sequence, verify you see "CA Connected" (CA: Channel Access) in the RunControl panel
- Press Button "cluster" in the GlobalControl panel to chose the correct data mode (mode 2: decomposed data, mode 3: raw data (traces)). If GT is not set up already for mode2 data taking (producing decomposed data) you should invoke option 9.
Note: In the Gretina RunControl panel, 'new dir' makes the run starts at 0002, switching back to an existing directory makes the run start at the previous Run number + 2
- Press button "global control"
The upcoming panel is being used for setting global parameters for ALL chrystals (baseline, polarity, etc.)
Raw delay: For internal mode data: 3.7 usec (for analyzing the traces)
typically you will see 2.7 usec here
Raw len: length of the trace in usec
don't go below 0.2 usec (corresponding to 20
short words, while the header is already 16 words long,
so there are 4 words left for the trace itself)
in decomposition mode: Use 2.0 usec
(corresponding to 16 words for the header and 184
words for the trace)
Threshold setting: typically cc: 1000, segment 300 (do not go below 300
in the segments and 150 in CC)
in "user trigger" you can set the crystal readout rate up to 20,000.
Lower this number if the decomposition cluster cannot keep up
- Press button "TimeStamps" in the Gretina DAQ top level to check if the timestamps are still synchronized imp sync: starts a train of imperative sync signals, by pressing the button again, the train stops single pulse imp sync: One single imperative sync pulse
- press button "pseudoScalers" in the Gretina DAQ top level to verify that data is coming in and being processed
- rebooting digitizer IOCs
ssh gsoper:[email protected] find the number in the "timestamp window". simply count to get the xx value to use. For crate 3, use 03. Do not ssh from the terminal you set up GT from as you will need to type "~." to get out and that will kill your window as well. Always ssh from a seperate terminal window
if you telnet into the IOC and you see tasks suspended (type i to see the processes and thir status) you need to reboot. Simply type "reboot". Using "tr" to start them usually does not help
after a reboot: push the "reset after an IOC reboot" butten and the "TTCS settings button" and the other buttons there.
for access to the trigger IOC,
ssh gsoper:[email protected]
Once all is set up, you can also start and stop runs on the command line with these commands (scripts in ~/scripts)
gDAQon (does not work?) gDAQoff (works)
The alarm page is accessed through the GT main DAQ screen upper right corner. This is where you can mute the alarm
to stop the DAQ
push the stop button in run control window
If you want to stop everything, select "K" in the $PWD/godaq script.
to shut down the DAQ
- Type 8 in the MENU to kill the Cluster
- Type K (Kill All) in the MENU to kill the rest
online sorting with GEBSort, to a map file
NOTE: you can only receive on-line data if the decomposition cluster is running. The Global Event Builder, where GEBSort gets it data from, is only active if the decomposition cluster is up and running
Here is an example of running GEBSort on ws3.gam taking data from the GEB to a map file we can look at while we take data
o ssh -X [email protected]
o Download [THIS IS OUT OF DATE]
cd workdir (e.g., /home/gtuser/gebsort) https://svn.anl.gov/repos/gs_analysis/GEBSort . or wget http://www.phy.anl.gov/gretina/GEBSort/AAAtar.tgz tar -zxvf AAAtar.tgz
on a1.gam you would need to do this as well
export ROOTSYS=/home/users/tl/root/root-v5-34-00 export PATH=$ROOTSYS/bin:$PATH export LD_LIBRARY_PATH=/home/users/tl/root/root-v5-34-00/lib/root
on ws3 or ws2 you should be fine
o now compile everything
rm curEPICS; ln -s /global/devel/base/R3.14.11 curEPICS make clean make make GEBSort ./mkMap > map.dat rm GTDATA; ln -s ./ GTDATA
o to run and take data, type
# +-- Global Event Builder (GEB) host IP (or simulator) # | + -- Number of events asked for on each read # | | +-- desired data type (0 is all) # | | | +-- timeout (sec) # | | | | ./GEBSort \ -input geb 10.0.1.100 100 0 100.0 \ -mapfile c1.map 200000000 0x9ef6e000 \ -chat GEBSort.chat
you may also just say
go geb_map
o to display the data in the map file
rootn.exe -l compile GSUtils (first line) sload c1.map file (third line) update (fourth line and other places) display
o note: It can take a while, up to a minute or so, before the GEB has data to offer to ./GEBSort so if ./GEBSort doesn't get data, just try again after you have started a run.
off-line sorting with GEBSort, to a root file
Here is an example of running GEBSort with output to a root file in an off-line situation
o Download the software
cd workdir (e.g., /home/gtuser/gebsort) git clone https://gitlab.phy.anl.gov/tlauritsen/GEBSort.git .
o now compile everything
make clean make ./mkMap > map.dat rm GTDATA; ln -s ./ GTDATA
Ignore errors about not being able to make 'GEBSort' if you don't have VxWorks and EPICS installed! It will make 'GEBSort_nogeb', which is the version of GEBSort we will use for off-line data sorting where VxWorks and EPICS are not needed (the on-line situation, where you DO need GEBSort, is described above).
o sort your (merged) file as:
./GEBSort_nogeb \ -input disk merged_data.gtd \ -rootfile test.root RECREATE \ -chat GEBSort.chat
note1: for DGS data, GEBSort_nogeb works equally well on the idividual files from gtReceiver4 as the merge of those files created with GEBMerge
o to display the data in the root file
rootn.exe -l (or 'root -l' since it is just a root file) compile GSUtils (first line) dload test.root file (third line) display
In general, the 'go' file of the GEBSort package contains a lot of examples. You can make your own script file to process your data using these examples.
FAQ
misc problem fixes and procedures
- To fix a non-responsive timestamp, i.e. just counts and never resets, use command "caput Cry[#]_CS_Fix (e.g., caput Cry4_CS_Fix 1) 1" where [#] is the bank # of the offending timestamp(s). This will reload the FPGA code and reboot the IOC. Any time an IOC is rebooted, you should press the "RESET after an IOC Reboot" button on Global Controls edm screen. Then choose the settings type you want, usually TTCS or Validate settings, on the same screen. Note: this only works if the IOC is still respnding to CA. If it doesn't, try access through the terminal server and reboot, or cycle the power on the crate. Typically you have to manually reboot when tExcTask and tNetTask are suspended.
- to select the decomposition nodes that should be runnin (and, thus, avoid any nodes that are broken), edit the files: "setup7modulesRaw.sh", "setupEndToEnd28.sh" in the directory: "/global/devel/gretTop/11-1/gretClust/bin/linux-x86". Then do 8 to kill , then run the scripts you just changed, and 6 to bring up decomposition cluster up again. On dogs, type 'wwtop', to see details of what nodes are running.
- to bring up the GT repair buttons: "~/gretinabuttons.tcl". you may have to do that from a1.gam for now as we have a problem doing some things from the linux WS machine;
- If some crystals are not getteing decomposed or running properly: stop, do imp sync, and start again. If that is not enough, also click on the 4 buttons in the south east corner of the "global Control" page. If it still does not decompose, stop the decomposition cluster and start it again in the main DAQ script
- set the buffer size in the GEB, use
caput GEB_CS_Delay 5
in this case, we set it to 5 seconds. 5 to 10 are good values.
- another mysterious thing to try
LEDAT 19 700;LEDAT 23 800;SetPZMultCC.sh;SETLEDWIN 30
Calibration Procedure ANL -- version
get the data
To obtain the data needed to do the segment and CC calibration, we change the GT configuration to
Raw Len = 0.20 us Trig Mode = Validate mode 3 data (in godaq script) the cluster nodes should be running 1 sendRaw process per crystal in cluster, select 'mode 2 and 3'
you may have to adjust 'Crystal readout Rate Limit' to adjust the rate. In this mode you can take data at a rate of 60 times faster than when you run your data through the decomposition cluster to obtain mode2 data. You can split the files using this handy utility
gMultiRun <number of runs> <minutes per run> <minutes between runs>
check that the
GlobalRaw.dat
file increments. There is also a Global.dat, but it should be empty.
run the calibration software
Shaofei is working on this software. Eventually we will add it to GEBSort, where the date will be processed in the 'bin_gtcal' module.
Calibration Procedure -- NSCL version
NOTE: these instructions were lifted from NSCL, we will have to modify them to the needs at ANL
In broad strokes, the procedure has been to fit source spectra for the central contacts (CC), and then calibrate the segments based on the observed energy in a segment vs. the calibrated CC energy for multiplicity 1 events. There are some simple, and fairly quick unpacking codes which make the necessary ROOT spectra, and then some ROOT macros that actually perform the fitting etc. The procedure is outlined below, and the codes are all located in /home/users/gretina/Calibration/.
CC Calibration
1. Unpack the calibration data (this all assumes it's mode3 data) using the code ./UnpackRaw, where the commands are
./UnpackRaw <Run#, i.e. 0061>
2. Run the macro to fit the peaks:
root -l
root [0] .L FitSpectra.C
root [1] FitCCSpectra(.root Filename, i.e. "MSUMay2012Run0060.root",
value for xmin, typically choose 100,
Boolean for manual checking, to check, set to 1, otherwise 0,
source type, i.e. "Eu152");
-- Here the inputs are the .root filename (i.e. "MSUMay2012Run0061.root") which was generated in the unpacking step, a minimum spectrum value to look at (not so important, just pick something low, like 100 or something), a boolean to say if you want to personally approve the fits (CHECK = 1), or just let it go (CHECK = 0), and the source type (options are "Co60", "Bi207", "Eu152", "Co56" and "Ra226" right now).
-- If you run this script with CHECK == 1, you run in a quasi-interactive mode. That is, for each CC, you'll get a chance to look at all of the fits, and to move to the next CC, you must accept the fits by hitting 'y', and then enter. If you see one you don't like, you can enter the peak number, and the fit will be redone, with tweaked parameters. Then you can either accept the peak, or reject it, in which case it's ignored in fitting the calibration. Peaks are numbered in order of increasing energy, and labeled in the ROOT canvas, so you know which number to type for a given fit. The only thing to note in this is that if peaks are < 20 keV apart, they are fit together. In this case, if peaks 7 + 8 are fit together, there will only be a spectrum shown in the position corresponding to peak 7. If you're unhappy with the fit though, and refit 7, it will only fit peak 7. If you want peak 8 retried as well, you need to specify this additionally.
-- Regardless of the value of CHECK, if the code can't find a peak, it'll ask you if you see one. If you do, you can enter the approximate channel number, and a fit will be attempted. If you don't, entering -1 will just leave out the calibration point.
-- Output from the macro:
paramCC<ROOTFilename>.dat (i.e. paramCCMSUMay2012Run0061.dat) -- contains all fit parameters meanCC<ROOTFilename>.dat (i.e. meanCCMSUMay2012Run0061.dat) -- contains just the centroid of the fits
3. After you have fit all the source spectra you're interested in, you can fit the calibration with the ROOT macro:
root -l
root [0] .L FitCalibration.C
root [1] FitCalibration(number of calibration files, i.e. 3,
First .root file, i.e. "MSUMay2012Run0060.root",
Second .root file, i.e. "MSUMay2012Run0071.root",
Third .root file, i.e. "MSUMay2012Run0070.root",
Fourth .root file, i.e. "" if only using three files,
Flag for CCONLY = 1 if only fitting CC,
Flag for making detector maps, DETMAPS = 0 for NO);
-- This literally fits the calibration points with a straight line, and calculates the residuals, etc. of the fit. Input to the macro is the number of files you want to use in the calibration (number), the .root filenames (i.e. "MSUMay2012Run0061.root"), a boolean to say if you only want to fit the CC (answer should be yes, so CCONLY = 1), and a boolean to see if you want to make the detector map files for decomposition (at this stage with only CC calibrations, no). Note, if you use less than 4 calibration files, for the unused filenames, just put "".
-- Output from the macro:
fitFile<ROOTFilename#1>.root (i.e. fitFileMSUMay2012Run0061.root) -- contains the calibration fit, and residual plots
finalCalOutput<ROOTFilename#1>.dat (i.e. finalCalOutputMSUMay2012Run0061.dat) -- contains three columns - ID, offset and slope This is the file I rename (EhiGainCor-CC-MMDDYYYY.dat) and use for CC calibration in unpacking codes.
finalDiffOutput<ROOTFilename#1>.dat (i.e. finalDiffOutputMSUMay2012Run0061.dat) -- contains residual information in text format
Segment Calibration
1. Edit the unpack-m1.C source code (in the subdirectory src/), and change the line that reads in the calibration to read the new CC calibration that you've created.
RdGeCalFile("<Name of CC calibration - i.e. finalFitFile<>.dat>", ehiGeOffset, ehiGeGain);
2. Remake the code, just type make. This replaces the executable in the directory above.
3. Run the code UnpackM1 on the data you're using for the segment calibration. Typically, pick something with statistics enough to see something in the back segments.
./UnpackMult1 <Directory, i.e. MSUMay2012> <Run Number, i.e. 0061>
-- This code takes a while, because it scans the file 28 times, to calibrate each crystal one at a time. This is because it's using 2D ROOT matrices, and I can't make the memory usage work properly to do multiple crystals at the same time. It always has a memory allocation problem. If someone knows how to fix this, by all means, please do. Let Heather know how you fix it too =)
-- The code is essentially sorting segment multiplicity 1 events, and using the calibrated CC to calibrate the segment energies. Right now, it's looking at the 5MeV CC to do this. You can change this in the code, if desired. At the end of the code, it then fits the plot of CC vs. segE, and extracts a slope and offset. This is actually then written out as a completed calibration file (see output information).
-- Output files from code:
<ROOTFilename>Crystal##.root where ## goes from 1 to 28 -- these are .root files containing the 2D plots for each crystal, as well as projections
<ROOTFilename>.slope -- contains channel ID (electronics ID), offset and slopes -- the finished calibration This is the file that is usually renamed to EhiGainCor-MMDDYYY.dat and used in codes.
Detector Map and Trace Gain Generation for Decomp
1. Edit the ROOT macro file FitCalibration.C, specifically the function MakeDetectorMaps(), to read in the appropriate calibration file. Also edit the destination directory for the generated detector map files, which appears in a few places...
calibrationFile = fopen("<CalibrationFilename.slope>", "r");
...
system("mkdir <DestinationDirectoryName>");
...
detmap_file = fopen(Form(<DestinationDirectoryName>/detmap_Q%dpos%d_CC%d.txt", quad, position, ccnum), "w");
...
tracegainfile = fopen(Form(<DestinationDirectoryName>/tr_gain_Q%dpos%d_CC%d.txt", quad, position, ccnum), "w");
2. Run the MakeDetectorMaps() macro.
root -l
root[0] .L FitCalibration.C
root[1] MakeDetectorMaps()
3. Detector map and trace gain files should be automatically generated in the directory specified within the macro.
Statistics required for calibration
* 56Co, mode 3, validate, trace 0.2us --- 8 GB = ~15 minutes * 152Eu, mode 3, validate, trace 0.2us --- 6 GB = ~15 minutes * 226Ra, mode 3, validate, trace 0.2us --- 8 GB = ~15 minutes
Statistics required for checking segments resolution
* 60Co (10 uC), mode 3, validate, trace 0.2us --- 30 GB = ~60 minutes --- 500 cts per segments of the rear layer