"How to" for analysis code: Difference between revisions
No edit summary |
No edit summary |
||
| (7 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
[last updated | [last updated Mar 28, 2022] | ||
Everything should be performed in digios/analysis/working directory, | Everything should be performed in '''digios/analysis/working''' directory, | ||
{| class="wikitable" | {| class="wikitable" | ||
| Line 15: | Line 15: | ||
|} | |} | ||
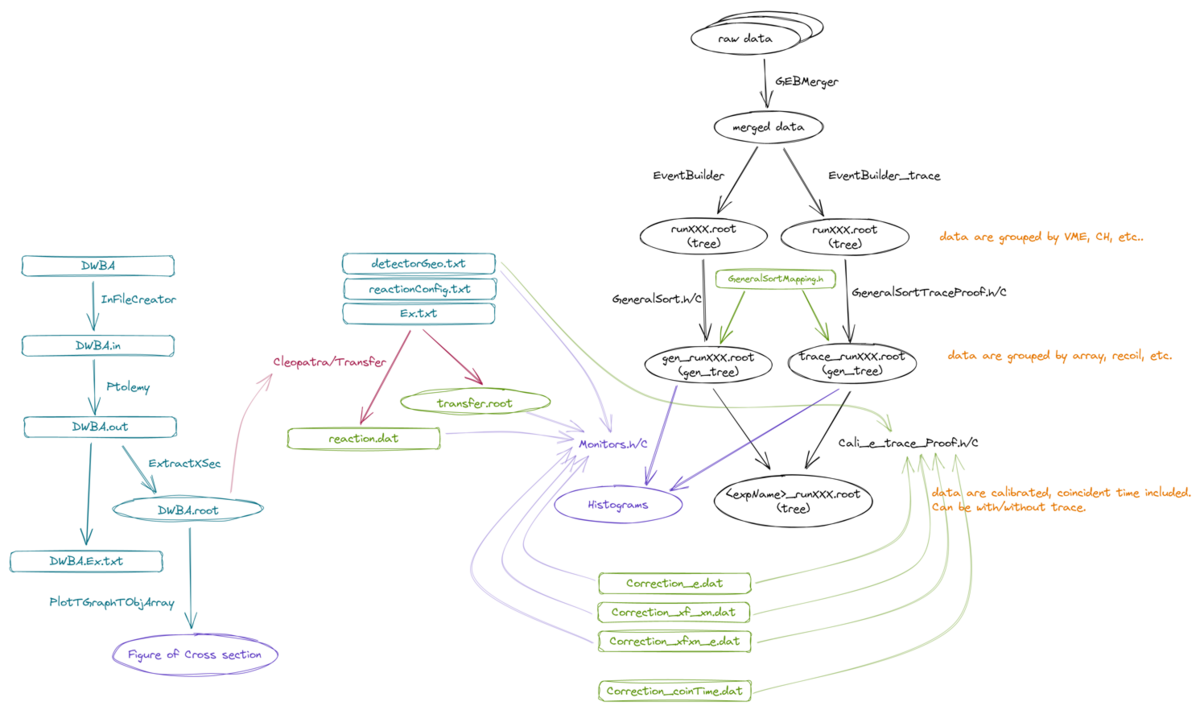
= | = Overview = | ||
[[File: | |||
[[File:Screenshot 2022-03-28 023801.png|1200px|frameless]] | |||
= General Usage = | = General Usage = | ||
| Line 47: | Line 48: | ||
Make sure you create folders or symbolic links for data, merged_data, and root_data under digios/analysis. | Make sure you create folders or symbolic links for data, merged_data, and root_data under digios/analysis. | ||
== How to update source code ? == | |||
The easiest easy is to restore everything that changed. | |||
digios>git reset --hard | |||
after resetting the folder, we can update the code by | |||
digios>git pull | |||
== How to get option for a method or function ? == | == How to get option for a method or function ? == | ||
Latest revision as of 19:29, April 29, 2022
[last updated Mar 28, 2022]
Everything should be performed in digios/analysis/working directory,
| ATTENTION: |
| txt file extension is editable for user. |
| dat files extension is not supposed to be edited, they are generated. |
| Editable file name will be highlighted with blue color |
Overview
General Usage
How to download the source code ?
make sure you have git in your computer.
in the directory you like to put in the code. for example, I like to put the code in ~/, then >cd ~/
~/>git clone https://github.com/calemhoffman/digios.git
you will see a folder name digios is there.
digios>cd digios
digios>git status
To check your experiment
digios>git branch -a
for example your experiment is using the branch h064_15N
digios>git status
you will see you are in the branch for your experiment.
for analysis, you only need the things inside digios/analysis
Make sure you create folders or symbolic links for data, merged_data, and root_data under digios/analysis.
How to update source code ?
The easiest easy is to restore everything that changed.
digios>git reset --hard
after resetting the folder, we can update the code by
digios>git pull
How to get option for a method or function ?
For most of the bash script, if you have no argument or the argument is "-help" it will display help message
working>Process_run
working>../Cleopatra/Transfer -help
For most root macro, for example, RDTCutsCreator(), after typed the first bracket, use tab, it will display the arguments.
root>RDTCutsCreator(
then press tab. it will show
root>RDTCutsCreator(TString filename, TString saveFileName = "rdtCuts.root", int eRange = 4000, int eRange = 60000 )
How to take a screenshot in MAC ?
Command + Shift + 4 = screenshot selected area
Command + Shift + 4, then space_bar = screenshot selected window
How to check the start time and stop time for a run?
in digios/analysis/data, there is RunTimeStamp.dat. this file stored all start and stop time for all runs.
in MAC or DAQ,
anywhere>ShowRunTimeStamp
How to check the raw files for a run?
in MAC or DAQ
anywhere>ShowRunSize [RUNNUM]
Git Branch Management
How to Setup/Switch for a new experiment?
| CAUTION: Advance user / user familiar with git only |
| CAUTION: Has to be done in the DAQ first |
| CAUTION: Has to be done in the MAC after |
given that the digios directory is clean, meaning
digios>git status
show no file is modified.
If you are not in the master branch
digios>./SetUpNewExp master
Then you are ready to setup a new experimental branch by
digios>./SetUpNewExp [new experiment name]
The bash script with do all the work.
After that, do the same in the MAC.
When the folder is not clean. i.e., not everything git committed. The SetUpNewExp will stop and not switch branches or make a new experiment.
In that case, the simplest way is
digios>git add -A # add all changes to stage digios>git commit -m "comment you like" # commit the change and make the folder clean
Or, you want to discard all change
digios>git reset --hard
How to clean up the working directory and prepare for a new experiment?
| CAUTION: Advance user / user familiar with git only |
| CAUTION: Need to be done in both MAC and DAQ |
| CAUTION: !!! READ till the end before you do anything!!!! |
After each experiment, the files in digios directory would be changed.
To check the change,
DAQ>cd ~/digios
digios>git status
To snapshot the change
digios>git add -A digios>git commit -m "comment for this snapshot"
It is better to push the snapshot to the github
digios>git push origin [branch name]
| CAUTION: Also need to git the DAQ! |
| CAUTION: The files in the DAQ and the MAC could be both changed. Mostly, the change in the DAQ is in digios/daq. |
| CAUTION: That means the DAQ and MAC has to be merged. |
We use a merge driver that the expName.sh will not be merged. The detail can be found in .gitattribute and git documentation.
It is better to start with the DAQ.
- commit and push the daq/edm/scripts folder
- commit and push the daq/edm/screens (if anything changed)
and then, go to the Mac,
- fetch and merge, since the DAQ pushed latest commit
- commit and push the analysis/Armory/
- commit and push the analysis/Cleopatra
- commit and push the analysis/working
Now, we can go to LCRC ( /lcrc/project/HELIOS/digios ) The LCRC always in master branch. then we can merge and solve conflict for each commit from the experiment.
After above action, we can go back to the DAQ, and commit the expName.sh, and switch back to master branch or setup a new experiment.
Process Run online
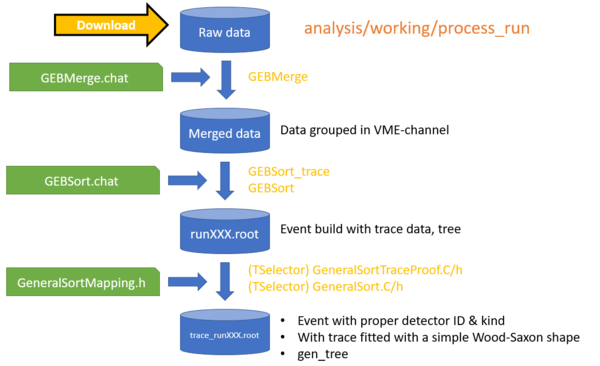
How to process run?
CAUTION: If the code is git clone or download recently, please go to ~/digios/analysis/GEBSort, and
GEBSort>make offline |
| CAUTION: In case of trace analysis, Please edit the GeneralSortMapping.h path in digios/analysis/Armory/GeneralSortTraceProof.C |
working>Process_RUN [RUNNUM]
The bash script will download run [RUNNUM], merge, event building, GeneralSort (mapping), and Monitors.C
to see more options, ./process_run without any argument
working>Process_RUN
Process_RUN [RunNum] [Merge] [EventBuild] [GeneralSort] [Monitor]"
RunNum = run number / \"lastRun\" "
Merge = 1/0/-1 || 1 = merge, 0 = not merge, -1 = force merge "
EventBld = 2/1/0/-1/-2 || 2 = with Trace"
GenralSort = n/1/0/-1/-n || 1 = GeneralSort.C, n = GeneralSortTraceProof.C with n-worker"
Monitors = 2/1/0 || 1 = single run, 2 = using the list in ChainMonitors.C"
10 = single run and post to websrv, 20 = list runs and post to websrv"
**** When feeding trace data to Monitors, the EventBld flag must be 2 or -2."
The working flow of process_run (without Monitor) is
For data calibration, see standard data analysis flow
How to process multiple runs at once ?
| CAUTION: In case of trace analysis, Please edit the GeneralSortMapping.h path in digios/analysis/Armory/GeneralSortTraceProof.C |
$./process_MultiRuns [RunNum1] [RunNum2] [Merge] [EventBuild] [GeneralSort]
RunNum1 = start run number
RunNum2 = stop run number
Merge = 1/0*
EventBld = 2/1/0* || 2 = with Trace
GenralSort = n/1/0* || 1 = GeneralSort.C, n = GeneralSortTraceProof.C with n-worker
* negative option = force
How to process multiple runs as a slurm job?
| CAUTION: In case of trace analysis, Please edit the GeneralSortMapping.h path in digios/analysis/Armory/GeneralSortTraceProof.C |
A slurm job is a job in LCRC. The script will generate the slurm.sh and submit the job to 1 node.
If only merge and build event, only 1 core will be used. If with GeneralSort, 36 cores will be used.
$./process_Slurm [RunNum1] [RunNum2] [time] [Merge] [EventBuild] [GeneralSort]
RunNum1 = start run number
RunNum2 = stop run number
time = allocate time
Merge = 1/0*
EventBld = 2/1/0* || 2 = with Trace
GenralSort = n/1/0* || 1 = GeneralSort.C, n = GeneralSortTraceProof.C with n-worker
* negative option = force
How to manually run the Monitors.C for a given run?
make sure gen_runXXX.root is generated in digios/analysis/root_data.
working>root ../root_data/gen_runXXX.root
working>gen_tree->Process("Monitors.C+")
How to "Monitor" multiple runs ?
edit ChainMonitors.C
you can see it use the TChain class.
to add files, for example, add the run 33 to 40, 42, 45, and 50, and I assume the run 33 to 40, 42, and 45 are already downloaded
chain->Add("../root_data/gen_run03[3-9].root");
chain->Add("../root_data/gen_run04[0,2,5].root");
chain->Add("../root_data/gen_run050.root");
Then, in process_run
working>./process_run 50 1 1 1 2
Notice that it will download, merge, build event, and GeneralSort (mapping) for run 50 ONLY.
How to change the array position ?
edit the file detectorGeo.txt
How to Donwload all runs into the Mac ?
working>Rsyn_data_from_DAQ.sh
How to change or add a recoil Cuts ?
see #How to use RDTCutsCreator ?
How to use RDTCutsCreator ?
After ./process_run, the ../Armory/RDTCutsCreator.C will be loaded.
root>RDTCutsCreator( [filename], [save file name], [E range], [dE range] )
The default [save file name] is "rdtCuts.root". If you like to use other [save file name], please correct the TString rdtCutFile in Monitors.C
if something go wrong during the RDTCutsCreator, simply delete the "rdtCuts.root".
There is a way to include multiple files for the RDTCutsCreator
root>RDTCutsCreator("../root_data/gen_run02[3-7].root")
or
root>RDTCutsCreator("../root_data/gen_run02[3,5,7,9].root")
To include more files, we can edit ../Armory/RDTCutsCreator.C
inside the file, manual add the files. To run that script.
working> root
root>.L ../Armory/RDTCutsCreator.C
root>RDTCutCreator("", [save file name], [E range], [dE range] )
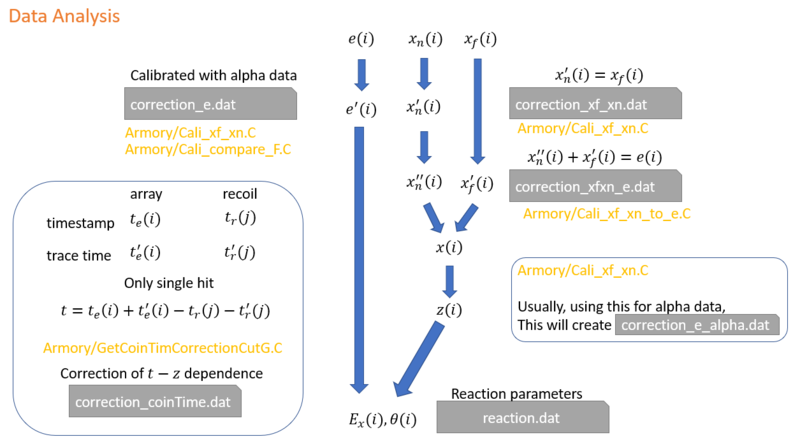
How is the Calibration flow ?
The calibration has few steps:
- Gain-matching with xn to xf
- Gain-matching with xn'+xf to e
- Calibrate e
To do step 1, It is better start with alpha data, because the alpha data is independent of position. By gating on 1 of the alpha peak, then plot the xf vs xn, the gain matching of xn to xf can be easily done. And because we need to gate on the alpha peak, then why not do a calibration on the energy using alpha data? So, in the Cali_xf_xn.C, the program will first calibrate alpha energy, then gate on 1 of the alpha peak to do gain match xn to xf.
To do step 2, we need a gain-matched xn, i.e. xn', so that xn'+xf should be proportional to the raw e.
In order to calibrate the energy, sometimes, alpha calibration is doing a good job, given that the alpha source is a good source. However, we can also calibrate the energy using kinematic E-Z lines. See How to Calibrate reaction data by kinematics ?
How to Calibrate alpha data ?
If statistics for single detector is sufficient, i.e. all peak is clearly identified.
edit the file ChainMonitors.C
working>root AutoCalibrationTrace.C+
select option 0
It will guide you through the process.
If statistics is not so good, we can use the ../Armory/Cali_e_single.C
How to Gain matching of xn to xf ?
working>root AutoCalibrationTrace.C+
select option 0
First, it will go through energy calibration, it does not need to be good.
Next, select any peak, so that the program will gate on that peak and plot xf vs xn. From there, the xn will gain match xf.
How to gain match the xn+xf to raw e ?
To properly do so, you have to gain-matched xn to xf. Then
working>root AutoCalibrationTrace.C+
select option 1
This has to be NOT alpha run, just any beam run. It will plot the raw e vs xf + xn'.
How to Calibrate reaction data by kinematics ?
( I know It needs more detail )
ONLY for sufficient statistic for individual detector.
edit the file ChainMonitors.C
working>root AutoCalibrationtrace.C+
select option 2
it will guide you through.
In order to speed up the process, it will create a temp.root. a root file only contain e and z.
User can use ../Armory/Check_e_x.root and create a cut to be used in the Monte-Carlo process.
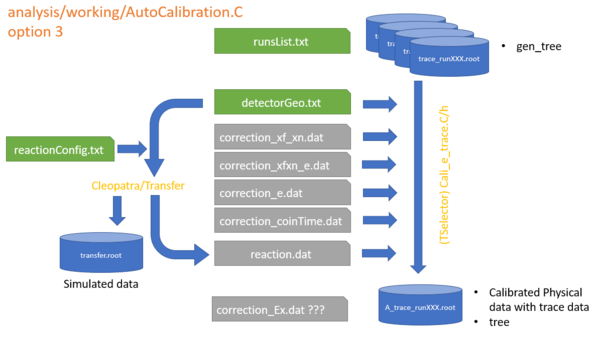
How to output a root file only contain physical data ?
after the calibration is done, we can edit ChainMonitors.C
and run
working>root AutoCalibrationTrace.C+
Select option 3 to generate the A_gen_runXXX.root
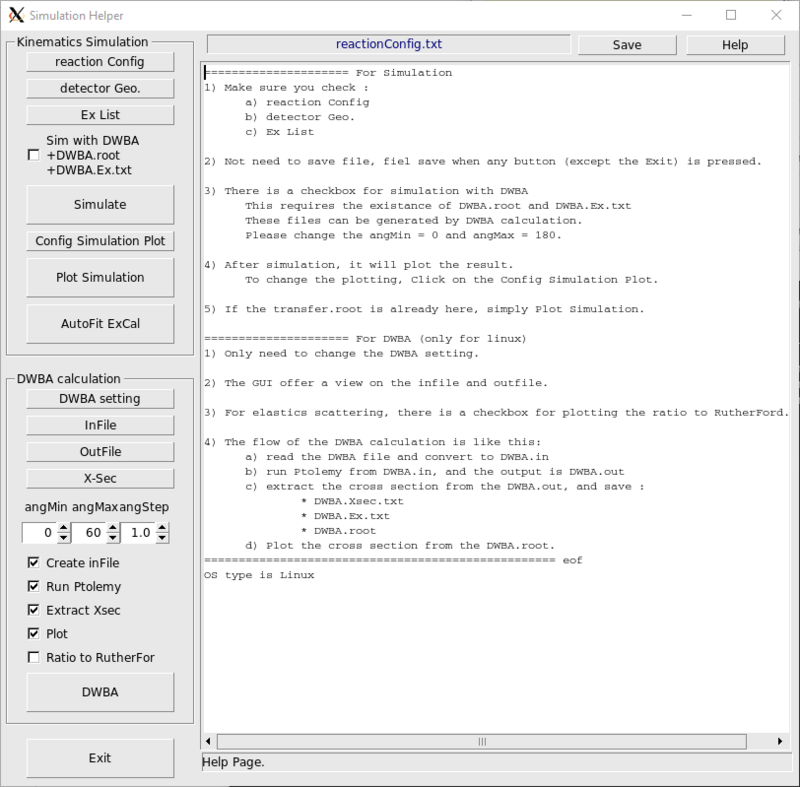
Kinematic and DWBA Calculation
The simplest way is using the Simulation_Helper.C. This is a GUI interface.
working>root Simulation_Helper.C
How to change the calculation for Ex and thetaCM ?
edit the file reactionConfig.txt, than recalculate the reaction.dat by running ../Cleopatra/Transfer
working>../Cleopatra/Transfer
How to change the kinematic E-Z line?
see #How to simulate the kinematics?
The E-Z lines are stored in fxList in transfer.root
In Monitors.C, at the end or Monitors::Terminate().
you can see the fxList.
How to simulate the kinematics?
CAUTION: If the code is git clone or download recently, please go to ~/digios/analysis/Cleopatra, and
Cleopatra>make |
edit
- reactionConfig.txt
- detectorGeo.txt
- Ex.txt
Then run
working>../Cleopatra/Transfer
after that, 2 files will be generated, 1) reaction.dat, 2) transfer.root
a tree is inside transfer.root.
you can see the tree branches by
root> tree->Print()
I hope the branch name is self-explained.
In Armory, there is a root macro Check_Transfer.C
working>root '../Armory/Check_Transfer.C("transfer.root")'
The macro load the transfer.root and plot the e-z plot, the recoil, recoil radius vs thetaCM etc...
How to do DWBA calculation ?
CAUTION: If the code is git clone or download recently, please go to ~/digios/analysis/Cleopatra, and
Cleopatra>make |
| CAUTION: ONLY IN LINUX, because Ptolemy only work in Linux. |
edit the file DWBA, then
working>./Cleopatra example 1 1 1 1 0 [minAng] [maxAng] [angStep]
This will generate the example.in, feed to Ptolemy, output example.out, extract the differential cross section into
- example.Xsec.txt
- example.root
And plot the d.s.c. using example.root
It will also generate example.Ex.txt
For arguments meaning, use Cleopatra without any argument
working>./Cleopatra
How to do kinematics simulation with DWBA differential cross section ?
see #How to do DWBA calculation ? first.
edit
- reactionConfig.txt
- detectorGeo.txt
- example
Make sure the reaction in the reactionConfig.txt is matching with that in example
working>./Cleopatra example 1 1 1 0 1 0 180 1
The excitation levels is using example.Ex.txt.
The bash script will calculate the DWBA d.s.c. using Cleopatra, then using ../Cleopatra/Transfer to do the simulation.
see #How to simulate the kinematics?
How to do trace analysis ?
updated Feb-12, 2021
simply
working>Process_RUN [RunNum] 1 -2 [number_of_worker] [Option_for_Monitors]
Updated Nov-6, 2019
The GEBSort_nogeb_trace was generated, Simple run ./process_run with option is fine
First, the GEBSort code has to be recompile by changing the file digios/GEBSort/bin_rcnp.C
in that file, comment out the trace (line ?)
Then, recompile
GEBSort>make offline
Now the gen_runXXX.root will contains trace data.
go back to working directory
working>./process_run [RUNNUM] 1 1 2 1
The 4th argument is 2, that means the GeneralSort (or the mapping ) will use ../Amrory/GeneralSortTrace.C instead of ../Amory/GeneralSort.C
The 4th argument can be also be more than 2, say, when it is N, it means it will use Proof, a parallel computing framework built-in in root that use N cores.
To display the trace, we can use ../Armory/readTrace.C ( this macro is kind of shitty.....)
How to count a 1D histogram for a given range ?
After ./process_run, a function Check1D() is loaded.
How to fit the excitation spectrum ?
If the reaction.dat is set properly, after ./process_run, the excite() function will give you the excitation spectrum.
root>excite()
You can use the fitAuto() function to auto fit the spectrum by
root>fitAuto(hEx)
For more option
fitAuto( [hist name], [BG estimation] )
Or you can can fit N-Gauss + 1 linear function. In this case, edit the AutFit_para.txt
root>fitNGaussP1(hEx)